Overview
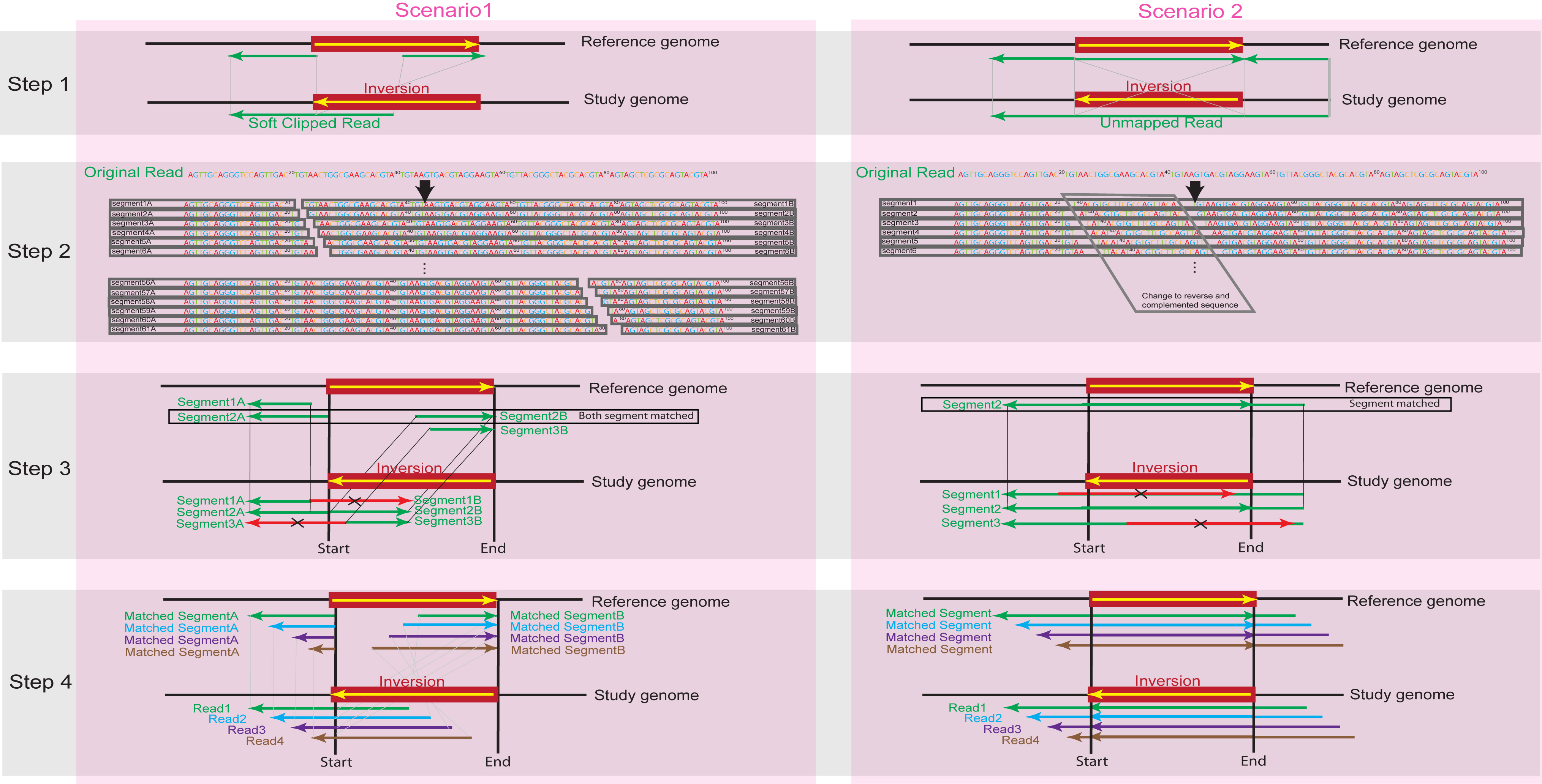
Above figure illustrates how SRinversion works in two different scenarios based on the different relationship between reads and the inversions
Method on detection of short inversions in human genome using next negeration sequencing data

Above figure illustrates how SRinversion works in two different scenarios based on the different relationship between reads and the inversions
SRinversion uses alignment results in bam format as input and mainly consists of 4 steps: 1) extract poorly mapped reads, 2) split or convert reads, 3) re-map reads, and 4) inversion region detection. For each step, two scenarios are considered based on different relationship between candidate reads and corresponding inversions