Professor CHUNG Hon Yin Brian (鍾侃言)

Clinical Associate Professor
-
MBBS(Hons, HKU), MSc(Genomics and Bioinformatics, CUHK), MD(HKU)
DCH(Ireland), MRCPCH (UK), FHKAM(Paediatrics), FRCPCH(UK), FCCMG(Clinical Genetics, Canada)
| bhychung@hku.hk
Tel: (852) 2255-4482
Fax: (852) 2855-1523
|
|
| Research ID ResearchGate Google Scholar ID (H-index: 46) ORCID ID Scopus ID HKU Scholars Hub |
|
Biography
Specialty
Clinical Genetics & Genomics
LinkedIn: www.linkedin.com/in/bhychung
Editorship or editorial board membership of scholarly journals
- 2012 - 2016: Secretary, Editorial Board, Hong Kong Journal of Paediatrics (HKJP)
- 2017 - now : Associate Editor, Editorial Board, Hong Kong Journal of Paediatrics (HKJP)
- 2015 - 2017: Member, Editorial Board, American Journal of Medical Genetics (Part A)
- 2018 - now : Associate Editor, Editorial Board, American Journal of Medical Genetics (Part A)
- 2019 - now : Associate Editor, Editorial board, npj Genomic Medicine
- 2019 - now : Associate Editor, Genetic Epidemiology Section, BMC Medical Genetics
- 2019 : Review Editor, American Journal of Medical Genetics (Part C) Seminar series – June 2019 issue on Clinical Genetics in Asia
- 2020 : Associate Editor, Editorial Board, Frontiers in Pediatrics
- 2020 : Associate Editor, Editorial Board, Frontiers in Genetics
- 2022 : Topic Editor, Genetics and Mechanism of Ciliopathies, Frontiers in Genetics
- 2021 - 2023: Associate Editor, Editorial Board, Journal of Translational Genetics and Genomics
- 2023 : Topic Editor, Genomics & Precision Health, Translational Genetics and Genomics
Professional Societies
- 2019 - now : President-Elect, Asia Pacific Society of Human Genetics
Awards
- Reviewers’ Choice - Shortlisted abstract for American Society of Human Genetics Annual Meeting on ‘Clinical use of the amniotic fluid cells transcriptome in deciphering Mendelian disease’ (2022)
- Gold Award (Team Award) at the QS Reimagine Education Award and the Teaching Innovation Award for Interprofessional education and collaborative practice (IPECP). Reimagine Education (2021)
- Team Award for interprofessional education and collaborative practice (IPECP) of the Teaching Excellence Awards, HKU (2021)
- Outstanding Teaching Award, Teaching Excellence Awards, HKU (2019) [ click here to open from uvision ]
- Best Paper Award - Teaching and Learning Physical Examination in the Clinical Setting: Authentic Assessment of Multi-domain Competencies for Independent Professional Practice; 2019 World Federation for Medical Education World Conference (shared with Dr Pamela Lee)
- Sir Patrick Manson Gold Medal (2018) - Clinical application of whole-genome technologies on Paediatric-onset rare diseases
- Faculty Teaching Medal (2018) [ click here to open related document ]
- Audience Award, Free paper, Frontiers in Medical and Health Sciences Education (2018, shared with Dr Pamela Lee)
- Award of Merit, Free paper, Frontiers in Medical and Health Sciences Education (2018, shared with Dr Pamela Lee)
- Best Young Investigator Prize - Hong Kong College of Paediatricians (2017)
- Knowledge Exchange Awards 2017 - Little People Care Alliance; Li Ka Shing Faculty of Medicine (2017, shared)
- Long Service Awards, After 15 Years of Service; Department of Paediatrics and Adolescent Medicine, Li Ka Shing Faculty of Medicine, The University of Hong Kong (2016)
- Outstanding Oral Presentation Award - Identifying genetic mutations in patients with RASopathies using a new generation sequencing diagnostic pipeline in Hong Kong; Annual Scientific Meeting 2013, Hong Kong College of Paediatricians (2013)
- Outstanding Poster Presentation Award - Integration of chromosomal microarray into paediatric clinical care in Hong Kong; Annual Scientific Meeting 2013, Hong Kong College of Paediatricians (2013)
- Certificate of Excellence - Master of Science in Genomics & Bioinformatics; Division of Genomics & Bioinformatics, CUHK-BGI Innovation Institute of Trans-omics (2013)
- Outstanding Team award - The Hong Kong West Cluster (2012)
- 10-year Loyalty Award - Queen Mary Hospital (2010)
- First Runner-up Best Poster Presentation - 7th Asia Pacific Medical Education Conference (APMEC) (2010)
- One of the Top 4 submissions - 34th Annual Scientific Meeting, Canadian College of Medical Genetics (2010)
- Best Basic Research - Annual Research Day, Paediatrics, Hospital for Sick Children, Toronto (2010)
- Fellow Award - 30th Annual David W. Smith Workshop on Malformations and Morphogenesis (2009 & 2010)
- Silver Medal in Best Original Research Contest - Clinical Markers Useful in Enhancing Diagnostic Yield for Children with Global Developmental Delay (GDD); HK Academy of Medicine (2005)
- Most Outstanding Free Paper - (SMARD1) Spinal muscular atrophy with respiratory distress type 1 mutation in a Chinese boy; The 3rd Hong Kong Genetic Symposium 2005 (2005)
Research Themes
My research focuses in three main areas:
- Medical application of whole genome technologies
- Clinical genetics & genetic counselling
- Multiomics & human diseases
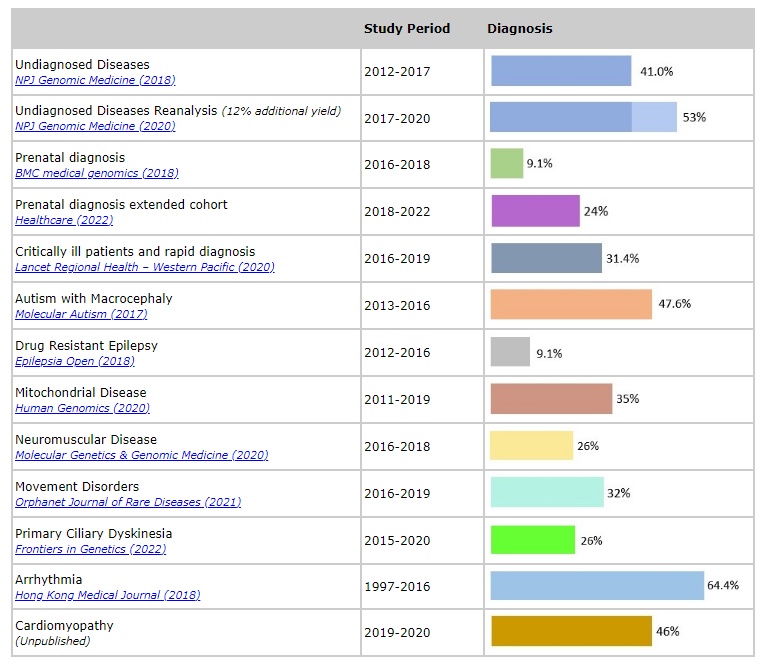
Genomic medicine is an emerging medical discipline that involves using genomic information about an individual as part of their clinical care and the health outcomes and policy implications of that clinical use. Its impact in clinical care depends significantly on the successful translation and integration of cutting-edge whole genome technologies from bench to bedside. My research interest is on the clinical application of whole genome technologies in the diagnosis of rare genetic diseases in children. My work includes a series of investigations using Chromosomal Microarray (CMA) in the genetic evaluation of congenital malformations, neurodevelopmental disorders, and congenital heart disease. I have also recruited over 1200 patients in the HKU paediatric exome sequencing project in various clinical contexts (see Figure).
To facilitate the implementation of genomic medicine in Hong Kong, we have also investigated our data further for implications in pre-emptive pharmacogenomics testing, expanded carrier screening, and incidental findings. From 2021, Dr. Chung servers as a Chief Scientific Officer of Hong Kong Genome Institute.
Clinical Genetics & Genetic Counselling
As a clinical geneticist, I have contributed to the advancement of diagnosis, management, and genetic counselling. This is achieved by syndrome delineation, phenotype expansion, characterization of founder mutations and providing insights for improving the management and better understanding of molecular mechanisms. Through leading a consortium of researchers from 60 institutes across 11 countries, we characterized 23 cases and discovered a new syndrome named MN1 C-terminal truncation (MCTT) syndrome. We have also discovered a novel role of the CC2D1A gene in heterotaxy and ciliary dysfunction.
Founder mutations are often responsible for the high prevalence of rare genetic disorders in specific populations and they are ethnicity-specific and typically under-represented in genomic databases. We identified founder mutations in the COQ4 gene for primary coenzyme Q10 deficiency-7, CFTR gene for Cystic Fibrosis, ALMS1 gene for Alstrom syndrome and KLHL40 gene for Nemaline Myopathy type 8.
Novel mutations discovered are archived on the online Clinvar database (https://www.ncbi.nlm.nih.gov/clinvar/submitters/507009/) hosted by the NCBI (National Center for Biotechnology Information, U.S. National Library of Medicine).
We have contributed several conditions of CTNBB1 in GeneReview and the translation of CTNNB1 genetics counselling reading materails.- Simplified Chinese Traditional Chinese
Multiomics and Human Disease
Multiomics is an approach to understand biological systems by a comprehensive, or global, assessment of a set of molecules simultaneously by high-throughput technologies. This includes genomics, epigenomics, transcriptomics, proteomics, metabolomics, which have given rise to the field of “integrative genetics” which integrates other technologies in the study of different molecular components in a biological system and how they contribute to human diseases across multiple ‘omics layers’.The use of multi-omics offers the opportunity to understand the flow of information and mechanism that underlies human disease , which can lead to an improvement in diagnosis, prevention and treatment strategies.
For example,Epigenetics is the study of heritable changes in gene expression caused by mechanisms other than changes in the underlying DNA sequence, and ‘epigenomics’ is the genome-wide study of such changes. Multiple studies have demonstrated the important role of epigenetics in interpreting genome data, especially in identifying regulatory elements that contain pathogenic non-coding mutations. My interest is in the study of epigenetics in rare (PIK3CA-Related Overgrowth Spectrum) and common (systemic lupus erythematosus (SLE)) diseases. I also described the first ever human case of paternal uniparental disomy for chromosome 19 using exome sequencing and DNA methylation array.
I am an active member of the International Epigene Consortium led by Prof Rosanna Weksberg of SickKids, Toronto with ongoing work in epigenetics. Our collaborative work has demonstrated disease-specific DNA methylation signature can be used to classify variants of uncertain significance, which was demonstrated in multiple genetic syndromes including Weaver syndrome and Sotos syndrome.
Another area of multiomics is ‘transcriptomics’ which involves the use of RNA sequencing (RNA-seq) to reveal the functional impact of genetic variants, especially variants of the non-coding genome on transcriptional changes, which cannot be revealed by genome sequencing (GS). Multiple studies have conducted RNA-seq on blood, muscle or skin fibroblast from patients with rare muscle or mitochondrial diseases for genetic diagnosis and raised the diagnostic yield by 10%-36% above ES/GS. In addition to postnatal diagnosis, RNA-seq was great potential in prenatal diagnostics. Recently, my team has conducted a proof-of-concept study titled “Diagnostic potential of the amniotic fluid cell transcriptome in deciphering Mendelian disease: a proof-of-concept”. We showed that clinically significant genes are well-expressed in in amniotic fluid cells, known as amniocyte, and their expression patterns are comparable to other clinically accessible tissues commonly used in RNA-seq study. As a proof-of-concept, 3 prenatal cases have been used for amniocyte RNA-seq which was analysed by our bioinformatic pipeline. We demonstrated that amniocyte RNA-seq data could provide strong functional evidence to reclassify variants for making diagnoses in fetuses with congenital anomalies. This study was selected as the “Reviewers’ Choice”, which is in the top 10% of poster abstracts in the American Society of Human Genetics (ASHG) Annual Meeting 2022 and published in NPJ Genomic in 2022.
Beyond the epigenetics of rare diseases, my research team is also interested in studying DNA methylation in common diseases, particularly systemic lupus erythematosus (SLE) as it is common in Chinese
Education
Dr Chung has served as the Education Committee Chairman of the Department 2017 - 2021 (co-chairman: Dr Pamela Lee).
He has served as the secretary of the Subspecialty Board of Genetics Genomics of the Hong Kong College of Paediatricians from 2018 - 2020. Now he is a member of the Subspecialty Board.
Dr Chung has achieved the status of Fellow (FHEA) in recognition of attainment against the UK Professional Standards Framework for teaching and learning support in higher education.
Teaching philosophy
I believe great teachers provide the environment and insights for the students to learn proactively and independently. I believe that clinical educators shall go beyond content knowledge and foster critical thinking and problem-solving skills (clinical reasoning). It is also particularly important to be clear about the relevance of the learning process to clinical practice. I take every opportunity to make use of real-life scenarios e.g. use of real patients or personal experience to enhance the student learning process. I strongly believe that learning should be patient-centered, as we are training future doctors to manage patients not only professionally but also humanely.
1) The learning process begins by identifying the gaps in the student's mindset, knowledge, and skills such that the student recognizes areas for development. Through effective feedback and techniques of self-reflection, our future doctors are equipped for their journey of self-long learning.
2) The learner must understand the relevance of the information in the clinical care of the patients. The use of real-life experiences in clinical practice is a powerful tool to engrave the relevance of the knowledge to their future practice as a doctor.
3) The ultimate aim of learning is not to be a good student but a good doctor, someone that can be entrusted with the important responsibilities of the health of Hong Kong citizens.
As a student (1995-1999) and teacher (2000-present) growing up in HKU, I embrace HKU’s vision to provide a “total learning experience”. The key institutional educational aims (https://tl.hku.hk/tl/) matches a lot of my core values in teaching: critical intellectual inquiry, life-long learning, critical self-reflection, collaboration, global citizenship and advocacy for the improvement of human conditions. In recognition of my contribution to teaching and curriculum development, I was awarded the Faculty Teaching Award in 2018-2019.
Teaching in Clinical Genetics
Click here to open the web resource for Teaching in Clinical Genetics ( *password needed to open this page )
Clinical Service
Selected publications
- Pei SLC, Chung BHY. Editorial: Genetics and mechanism of ciliopathies. Front Genet. 2022 Oct 28; 13:1067168. Link
- Chau JFT, Lee M, Chui MMC, Yu MHC, Fung JLF, Mak CCY, Chau CS-K, Siu KK, Hung J, Yeung KS, Kwong AKY, O'Callaghan C, Lau YL, Lee C-WD, Chung BHY and Lee S-L (2022) Functional Evaluation and Genetic Landscape of Children and Young Adults Referred for Assessment of Bronchiectasis. Front Genet. 2022 Jul 19. (IF 4.772) Link
- Biesecker LG, Adam MP, Chung BHY, Kosaki K, Menke LA, White SM, Carey JC, Hennekam RCM. Elements of morphology: Standard terminology for the trunk and limbs. American Journal of Medical Genetics (Part A) 2022 Nov;188(11):3191-3228. (IF 2.578) Link
- Chau JFT, Yu MHC, Chui MMC, Yeung CCW, Kwok AWC, Zhuang X, Lee R, Fung JLF, Lee M, Mak CCY, Ng NYT, Chung CCY, Chan MCY, Tsang MHY, Chan JCK, Chan KYK, Kan ASY, Chung PHY, Yang W, Lee SL, Chan GCF, Tam PKH, Lau YL, Yeung KS, Chung BHY, Tang CSM. Comprehensive analysis of recessive carrier status using exome and genome sequencing data in 1543 Southern Chinese. NPJ Genom Med. 2022 Mar 21. (IF 8.617) Link
- Yu MHC, Chan MCY, Chung CCY, Li AWT, Yip CYW, Mak CCY, Chau JFT, Lee M, Fung JLF, Tsang MHY, Chan JCK, Wong WHS, Yang J, Chui WCM, Chung PHY, Yang W, Lee SL, Chan GCF, Tam PKH, Lau YL, Tang CSM, Yeung KS, Chung BHY. Actionable pharmacogenetic variants in Hong Kong Chinese exome sequencing data and projected prescription impact in the Hong Kong population. PLoS Genet. 2021 Feb 18;17(2):e1009323. (IF 5.917) Link
- Chung BHY, Chau JFT, Wong GK. Rare versus common diseases: a false dichotomy in precision medicine. NPJ Genom Med. 2021;6(1):19. Published 2021 Feb 24. doi:10.1038/s41525-021-00176-x (IF 8.617) Link
- Yu MHC, Chau JFT, Au SLK, Lo HM, Yeung KS, Fung JLF, Mak CCY, Chung CCY, Chan KYK, Chung BHY, Kan ASY. Evaluating the Clinical Utility of Genome Sequencing for Cytogenetically Balanced Chromosomal Abnormalities in Prenatal Diagnosis. Front Genet. 2021 Jan 27;11:620162. (IF 4.772) Link
- Ma ACH, Mak CCY, Yeung KS, Pei SLC, Ying D, Yu MHC, Hasan KMM, Chen X, Chow PC, Cheung YF, Chung BHY. Monoallelic Mutations in CC2D1A Suggest a Novel Role in Human Heterotaxy and Ciliary Dysfunction. Circulation: Precision and Genomic Medicine. 2020 Dec;13(6):e003000. (IF 7.465) Link
- Yu MHC, Mak CCY, Fung JLF, Lee M, Tsang MHY, Chau JFT, Chung PH, Yang W, Chan GCF, Lee SL, Lau YL, Tam PKH, Tang CSM, Yeung KS, Chung BHY. Actionable secondary findings in 1116 Hong Kong Chinese based on exome sequencing data. J Hum Genet. 2020 Nov 22:1-5. Link (IF 3.755)
- Chau JFT, Chung CCY, Yu MHC, Yeung KS, Chung BHY. “Rare” Monogenic Variants in Severe COVID-19 cases. Science. (e-Letter, 2020 Nov 19) Link (IF 47.728)
- Chung CCY, Leung GKC, Mak CCY, Fung JLF… Lau YL, Chan GCF, Lee SL, Yeung KS, Chung BHY. Rapid whole-exome sequencing facilitates precision medicine in paediatric rare disease patients and reduces healthcare costs. The Lancet Regional Health-Western Pacific. 2020 Aug 1;1:100001. Link (IF 8.559)
- Mak CCY, Doherty D, Lin AE, Vegas N, Cho MT, Viot G, Dimartino C, Weisfeld-Adams JD, Lessel D, Joss S, Li C, Gonzaga-Jauregui C, Zarate YA, Ehmke N, Horn D, Troyer C, Kant SG, Lee Y, Ishak GE, Leung G, Barone Pritchard A, Yang S, Bend EG, Filippini F, Roadhouse C, Lebrun N, Mehaffey MG, Martin PM, Apple B, Millan F, Puk O, Hoffer MJV, Henderson LB, McGowan R, Wentzensen IM, Pei S, Zahir FR, Yu M, Gibson WT, Seman A, Steeves M, Murrell JR, Luettgen S, Francisco E, Strom TM, Amlie-Wolf L, Kaindl AM, Wilson WG, Halbach S, Basel-Salmon L, Lev-El N, Denecke J, Vissers LELM, Radtke K, Chelly J, Zackai E, Friedman JM, Bamshad MJ, Nickerson DA; University of Washington Center for Mendelian Genomics, Reid RR, Devriendt K, Chae JH, Stolerman E, McDougall C, Powis Z, Bienvenu T, Tan TY, Orenstein N, Dobyns WB, Shieh JT, Choi M, Waggoner D, Gripp KW, Parker MJ, Stoler J, Lyonnet S, Cormier-Daire V, Viskochil D, Hoffman TL, Amiel J, Chung BHY*, Gordon CT*. MN1 C-terminal truncation syndrome is a novel neurodevelopmental and craniofacial disorder with partial rhombencephalosynapsis. Brain. 2020;143(1):55-68. Link (IF 15.255) *Co-corresponding Author
- Yu MHC, Tsang MHY, Lai S, Ho MSP, Tse DML, Wills B, Kwon gAKY, Chou YY, Lin SP, Quinzli CM, Hwu WL, Chien YH, Kuo PL, Chan VCM, Tsoi C,, Chong SC, Rodenburg RJT, Smeitink J, Mak CCY, Yeung KS, Fung JLF, Lam W, Hui J, Lee NC, Fung CW, Chung BHY. Primary coenzyme Q10 deficiency-7: expanded phenotypic spectrum and a founder mutation in southern Chinese. NPJ Genomic Medicine 2019; 4:18. Link (IF 8.617)
- Zayts O, Shipman H, Fung JLF, Liu APY, Kwok SY, Tsai ACH, Yung TC, Chung BHY. The different facets of "culture" in genetic counseling: A situated analysis of genetic counseling in Hong Kong. Am J Med Genet C Semin Med Genet. 2019;181(2):187-195. Link (IF 3.359)
- Chiu ATG, Chung CCY, Wong WHS, Lee SL, Chung BHY. Healthcare burden of rare diseases in Hong Kong–adopting ORPHAcodes in ICD-10 based healthcare administrative datasets. Orphanet Journal of Rare Diseases. 2018 Dec 1;13(1):147. Link (IF 4.303)
- Yeung KS, Ho MSP, Lee SL, Kan ASY, Chan KYK, Tang MHY, Mak CCY, Leung GKC, So PL, Pfundt R, Marshall CR, Scherer SW, Choufani S, Weksberg R, Chung BHY. Paternal uniparental disomy of chromosome 19 in a pair of monochorionic diamniotic twins with dysmorphic features and developmental delay. Journal of Medical Genetics 2018;55(12):847-852 Link (IF 5.941)
- Mak CCY, Leung GKC, Mok GTK, Yeung KS, Yang W, Fung CW, Chan SHS, Lee SL, Lee NC, Pfundt R, Lau YL, Chung BHY. Exome sequencing for paediatric-onset diseases: impact of the extensive involvement of medical geneticists in the diagnostic odyssey. NPJ Genom Med. 2018;3:19. Link (IF 8.617)
- Yeung KS, Tso WWY, Ip JJK, Mak CCY, Leung GKC, Tsang MHY, Ying D, Pei SLC, Lee SL, Yang W, Chung BHY. Identification of mutations in the PI3K-AKT-mTOR signalling pathway in patients with macrocephaly and developmental delay and/or autism. Molecular Autism 2017; 8:66.Link (IF 6.476)
- Mak ASL, Chiu ATG, Leung GKC, Mak CCY, Chu YWY, Mok GTK, Tang WF, Chan KYK, Tang MHY, Lau ETK, So KW, Tao VQ, Fung CW, Wong VCN, Uddin M, Lee SL, Marshall CR, Scherer SW, Kan ASY, Chung BHY. Use of clinical chromosomal microarray in Chinese patients with autism spectrum disorder-implications of a copy number variation involving DPP10. Molecular Autism. 2017;8:31. Link (IF 6.476)
- Choufani, S, Cytrynbaum C, Chung BHY, Turinsky A, Grafodatskaya D, Chen YA, Cohen A, Dupuis L, Butcher D, Siu MT, Luk HM, Lo I, Lam S, Caluseriu O, Stavropoulos D, Reardon W, Mendoza-Londono R, Brudno M, Gibson W, Chitayat D, Weksberg R. NSD1 mutations generate a genome-wide DNA methylation signature. Nature Communications 2015;6:10207. Link(IF 17.694)







